
Data Wrangling - one data frame 🛠
MATH/COSC 3570 Introduction to Data Science
Department of Mathematical and Statistical Sciences
Marquette University
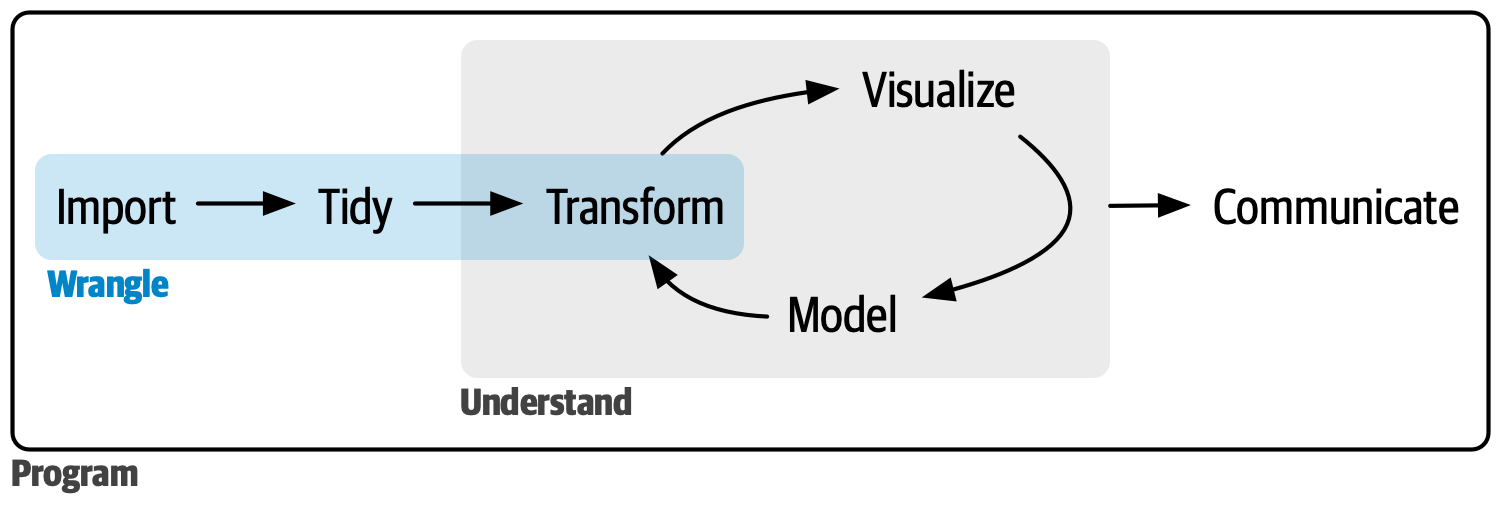
Grammar of Data Manipulation
Grammar of Data Wrangling: dplyr 📦
- based on the concepts of functions as verbs that manipulate data frames
-
mutate: create new columns from the existing1 -
filter: pick rows matching criteria -
slice: pick rows using index(es) -
distinct: filter for unique rows -
select: pick columns by name -
summarise: reduce variables to values -
group_by: for grouped operations -
arrange: reorder rows - … (many more)
Rules of dplyr Functions
First argument is always a data frame
Subsequent arguments say what to do with that data frame
Always return a data frame
Don’t modify in place
Data: US gun murders by state for 2010
# A tibble: 51 × 5
state abb region population total
<chr> <chr> <chr> <dbl> <dbl>
1 Alabama AL South 4779736 135
2 Alaska AK West 710231 19
3 Arizona AZ West 6392017 232
4 Arkansas AR South 2915918 93
5 California CA West 37253956 1257
6 Colorado CO West 5029196 65
# ℹ 45 more rowsAdding a New Variable (Column) with mutate()
-
dplyr::mutate()takes- a data frame as the 1st argument
- the name and values of the variable as the 2nd argument using format
name = values.
# A tibble: 51 × 6
state abb region population total rate
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Alabama AL South 4779736 135 2.82
2 Alaska AK West 710231 19 2.68
3 Arizona AZ West 6392017 232 3.63
4 Arkansas AR South 2915918 93 3.19
5 California CA West 37253956 1257 3.37
6 Colorado CO West 5029196 65 1.29
# ℹ 45 more rowstotalandpopulationinside the function are not defined in our R environment.dplyrfunctions know to look for variables in the data frame provided in the 1st argument.
Filtering Observations (Rows) with filter()
-
dplyr::filter()takes a- data frame as the 1st argument
- conditional statement as the 2nd. (pick rows matching criteria)
# filter the data table to only show the entries for which
# the murder rate is lower than 0.7
murders |>
filter(rate < 0.7) #<<# A tibble: 5 × 6
state abb region population total rate
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Hawaii HI West 1360301 7 0.515
2 Iowa IA North Central 3046355 21 0.689
3 New Hampshire NH Northeast 1316470 5 0.380
4 North Dakota ND North Central 672591 4 0.595
5 Vermont VT Northeast 625741 2 0.320
filter() for Many Conditions at Once
Logical Operators
| operator | definition | operator | definition |
|---|---|---|---|
< |
less than |
x | y
|
x OR y
|
<= |
less than or equal to | is.na(x) |
if x is NA
|
> |
greater than | !is.na(x) |
if x is not NA
|
>= |
greater than or equal to | x %in% y |
if x is in y
|
== |
exactly equal to | !(x %in% y) |
if x is not in y
|
!= |
not equal to | !x |
not x
|
x & y |
x AND y
|
slice() for Certain Rows using Indexes
# A tibble: 4 × 6
state abb region population total rate
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Arizona AZ West 6392017 232 3.63
2 Arkansas AR South 2915918 93 3.19
3 California CA West 37253956 1257 3.37
4 Colorado CO West 5029196 65 1.29How do we subset rows using matrix indexing?
distinct() to Filter for Unique Rows
# A tibble: 4 × 1
region
<chr>
1 South
2 West
3 Northeast
4 North Central# A tibble: 4 × 6
state abb region population total rate
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Alabama AL South 4779736 135 2.82
2 Alaska AK West 710231 19 2.68
3 Connecticut CT Northeast 3574097 97 2.71
4 Illinois IL North Central 12830632 364 2.84
distinct() Grabs First Row of The Unique Value
# A tibble: 4 × 6
state abb region population total rate
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Alabama AL South 4779736 135 2.82
2 Alaska AK West 710231 19 2.68
3 Connecticut CT Northeast 3574097 97 2.71
4 Illinois IL North Central 12830632 364 2.84Selecting Columns with select()
- In
dplyr::select(), the 1st argument is a data frame, followed by variable names being selected in the data. - The order of variable names matters!
select() to Exclude Variables
# A tibble: 51 × 5
state abb region total rate
<chr> <chr> <chr> <dbl> <dbl>
1 Alabama AL South 135 2.82
2 Alaska AK West 19 2.68
3 Arizona AZ West 232 3.63
4 Arkansas AR South 93 3.19
5 California CA West 1257 3.37
6 Colorado CO West 65 1.29
# ℹ 45 more rows
select() a Range of Variables
[1] "state" "abb" "region" "population" "total"
[6] "rate" # A tibble: 51 × 4
region population total rate
<chr> <dbl> <dbl> <dbl>
1 South 4779736 135 2.82
2 West 710231 19 2.68
3 West 6392017 232 3.63
4 South 2915918 93 3.19
5 West 37253956 1257 3.37
6 West 5029196 65 1.29
# ℹ 45 more rows
select() Variables with Certain Characteristics
-
starts_with()is a tidy-select helper function.
select() Variables with Certain Characteristics
-
ends_with()is a tidy-select helper function.
tidy-select Helpers1
-
starts_with(): Starts with a prefix -
ends_with(): Ends with a suffix -
contains(): Contains a literal string -
num_range(): Matches a numerical range like x01, x02, x03 -
one_of(): Matches variable names in a character vector -
everything(): Matches all variables -
last_col(): Select last variable, possibly with an offset -
matches(): Matches a regular expression (a sequence of symbols/characters expressing a string/pattern to be searched for within text)
Rationale for Pipe Operator
How do we show three variables (state, region, rate) for states that have murder rates below 0.7?
- Method 1: Define the intermediate object
new_table
Rationale for Pipe Operator
How do we show three variables (state, region, rate) for states that have murder rates below 0.7?
- Method 2: Apply one function onto the other with no intermediate object
Rationale for Pipe Operator
- The code that looks like a verbal description of what we want to do without intermediate objects:
data > select() > data after selecting > filter() > data after selecting and filtering
Summarizing Data – summarize()
-
summarize()provides a data frame that summarizes the statistics we compute.
Summarizing Data – summarize()
(s <- heights |>
filter(sex == "Female") |>
summarize(avg = mean(height),
stdev = sd(height),
median = median(height),
minimum = min(height)))# A tibble: 1 × 4
avg stdev median minimum
<dbl> <dbl> <dbl> <dbl>
1 64.9 3.76 65.0 51[1] 64.9[1] 51summarise() produces a new data frame that is not any variant of the original data frame.
Summarizing Data – summarize()
- One variable
quansthat has 3 values. The output is a 3 by 1 data frame.
Grouping – group_by()
# A tibble: 1,050 × 2
# Groups: sex [2]
sex height
<chr> <dbl>
1 Male 75
2 Male 70
3 Male 68
4 Male 74
5 Male 61
6 Female 65
# ℹ 1,044 more rowsheights_groupis a grouped data frame.Tibbles are similar, but see
Groups: sex [2]after grouping data bysex.summarize()behaves differently when acting ongrouped_df.
Group and Summarize: group_by() + summarize()
-
summarize()applies the summarization to each group separately.
Sorting Rows in Data Frames – arrange()
-
arrange()orders entire data tables.
# A tibble: 51 × 6
state abb region population total rate
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Wyoming WY West 563626 5 0.887
2 District of Columbia DC South 601723 99 16.5
3 Vermont VT Northeast 625741 2 0.320
4 North Dakota ND North Central 672591 4 0.595
5 Alaska AK West 710231 19 2.68
6 South Dakota SD North Central 814180 8 0.983
# ℹ 45 more rows15-dplyr
In lab.qmd ## Lab 15 section, import the murders.csv data and
Add (mutate) the variable
rate = total / population * 100000tomurdersdata (as I did).Filter states that are in region Northeast or West and their murder rate is less than 1.
Select variables
state,region,rate.
Print the output table after you do 1. to 3., and save it as object
my_states.Group
my_statesbyregion. Then summarize data by creating variablesavgandstdevthat compute the mean and standard deviation ofrate.Arrange the summarized table by
avg.
state region rate
1 Hawaii West 0.515
2 Idaho West 0.766
3 Maine Northeast 0.828
4 New Hampshire Northeast 0.380
5 Oregon West 0.940
6 Utah West 0.796
7 Vermont Northeast 0.320
8 Wyoming West 0.887# A tibble: 2 × 3
region avg std_dev
<fct> <dbl> <dbl>
1 Northeast 0.509 0.278
2 West 0.781 0.164Data Manipulation
New Variables .assign
Have to use
murders.totalandmurders.populationinstead oftotalandpopution.
Filter Rows .query
Conditions must be a string to be evaluated!
Cannot write
murders.rate, and should userate.
Select Columns .filter
Have to be strings
region rate state
0 South 2.82 Alabama
1 West 2.68 Alaska
2 West 3.63 Arizona
3 South 3.19 Arkansas
4 West 3.37 California
5 West 1.29 Colorado
6 Northeast 2.71 Connecticut
7 South 4.23 Delaware
8 South 16.45 District of Columbia
9 South 3.40 Florida
10 South 3.79 Georgia
11 West 0.51 Hawaii
12 West 0.77 Idaho
13 North Central 2.84 Illinois
14 North Central 2.19 Indiana
15 North Central 0.69 Iowa
16 North Central 2.21 Kansas
17 South 2.67 Kentucky
18 South 7.74 Louisiana
19 Northeast 0.83 Maine
20 South 5.07 Maryland
21 Northeast 1.80 Massachusetts
22 North Central 4.18 Michigan
23 North Central 1.00 Minnesota
24 South 4.04 Mississippi
25 North Central 5.36 Missouri
26 West 1.21 Montana
27 North Central 1.75 Nebraska
28 West 3.11 Nevada
29 Northeast 0.38 New Hampshire
30 Northeast 2.80 New Jersey
31 West 3.25 New Mexico
32 Northeast 2.67 New York
33 South 3.00 North Carolina
34 North Central 0.59 North Dakota
35 North Central 2.69 Ohio
36 South 2.96 Oklahoma
37 West 0.94 Oregon
38 Northeast 3.60 Pennsylvania
39 Northeast 1.52 Rhode Island
40 South 4.48 South Carolina
41 North Central 0.98 South Dakota
42 South 3.45 Tennessee
43 South 3.20 Texas
44 West 0.80 Utah
45 Northeast 0.32 Vermont
46 South 3.12 Virginia
47 West 1.38 Washington
48 South 1.46 West Virginia
49 North Central 1.71 Wisconsin
50 West 0.89 WyomingGrouping .groupby + .agg
dplyr::group_by() + dplyr::summarize()
Sorting .sort_values
state abb region population total rate
50 Wyoming WY West 563626 5 0.89
8 District of Columbia DC South 601723 99 16.45
45 Vermont VT Northeast 625741 2 0.32
34 North Dakota ND North Central 672591 4 0.59
1 Alaska AK West 710231 19 2.68dplyr::arrange(desc())
state abb region population total rate
8 District of Columbia DC South 601723 99 16.45
18 Louisiana LA South 4533372 351 7.74
25 Missouri MO North Central 5988927 321 5.36
20 Maryland MD South 5773552 293 5.07
40 South Carolina SC South 4625364 207 4.48