Probability and Statistics 🎲
MATH/COSC 3570 Introduction to Data Science
Department of Mathematical and Statistical Sciences
Marquette University
Why Study Probability
- We live in a world full of chances and uncertainty!
Mar 29, 2022
Mar 22, 2023
Mar 18, 2024



Why Study Probability
Probability is the study of chance, the language of uncertainty.
-
We could do data science without any probability involved. However, what we can learn from data will be much limited. Why?
Every time you collect a data set, you obtain a different one.
Your data are affected by some chance or random noises!
Knowledge of probability is essential for data science, especially when we want to quantify uncertainty about what we learn from our data.
Probability as Relative Frequency
The probability that some outcome of a process will be obtained is the relative frequency with which that outcome would be obtained if the process were repeated a large number of times independently under similar conditions.
-
Example:
- toss a coin: probability of getting heads 🪙
- pick a ball (red/blue) in an urn: probability of getting a red ball 🔴 🔵
Frequency Relative_Frequency
Heads 6 0.6
Tails 4 0.4
Total 10 1.0
---------------------
Frequency Relative_Frequency
Heads 535 0.54
Tails 465 0.47
Total 1000 1.00
---------------------Monte Carlo Simulation for Categorical Data
- We get a bag of 5 balls colored in red or blue.
Without peeking at the bag, how do we approximate the probability of getting a red ball?

Monte Carlo Simulation: Repeat drawing a ball at random a large number of times to approximate the probability by the relative frequency of getting a red ball.
So how many red balls in the bag?
Random Seed set.seed()
- When doing sampling, we use random number generators, and results vary from sample to sample.
- To ensure the results are the same every time we do sampling, set the random seed to a specific number by
set.seed()
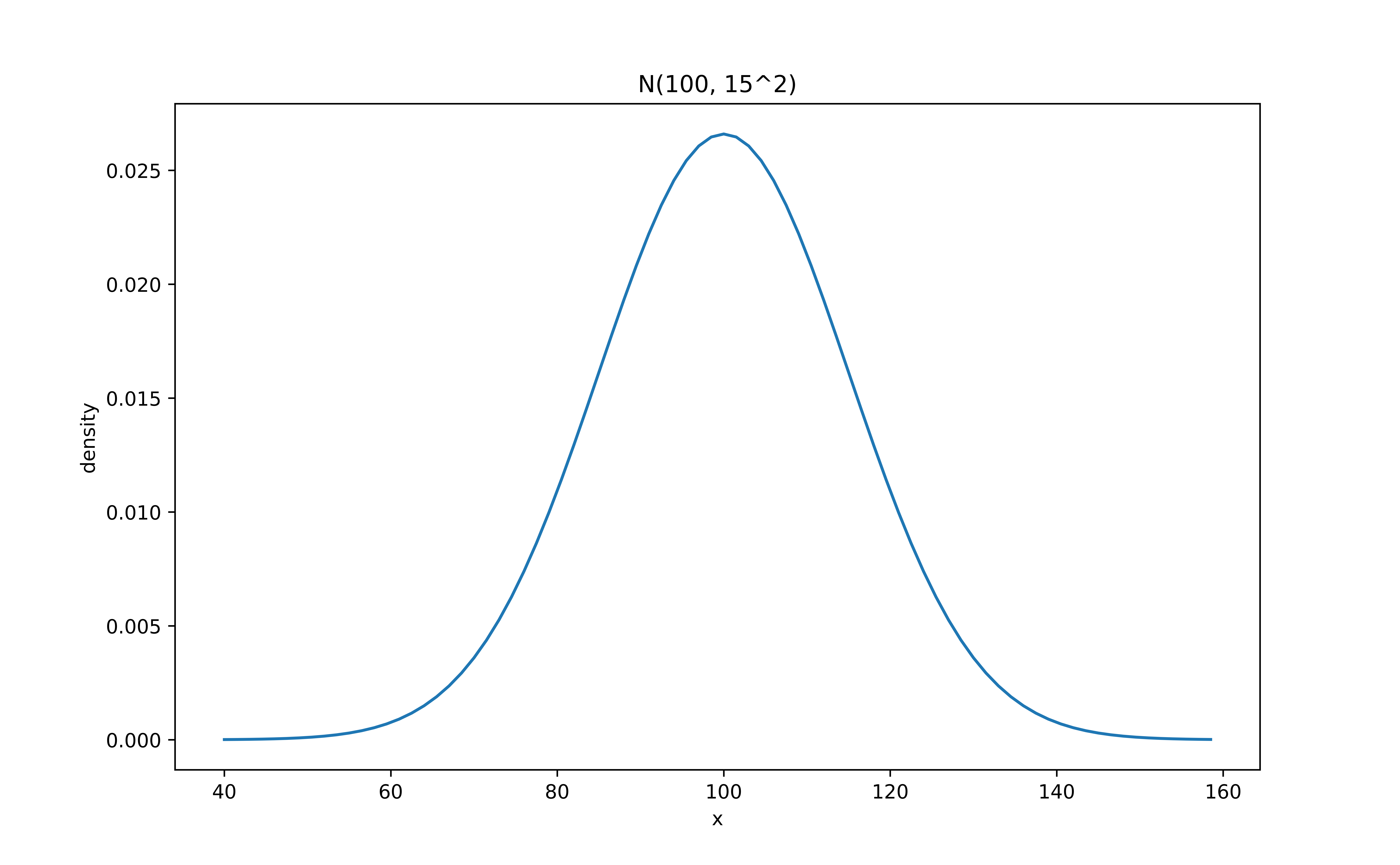
Normal (Gaussian) Distribution \(N(\mu, \sigma^2)\)
- Density curve
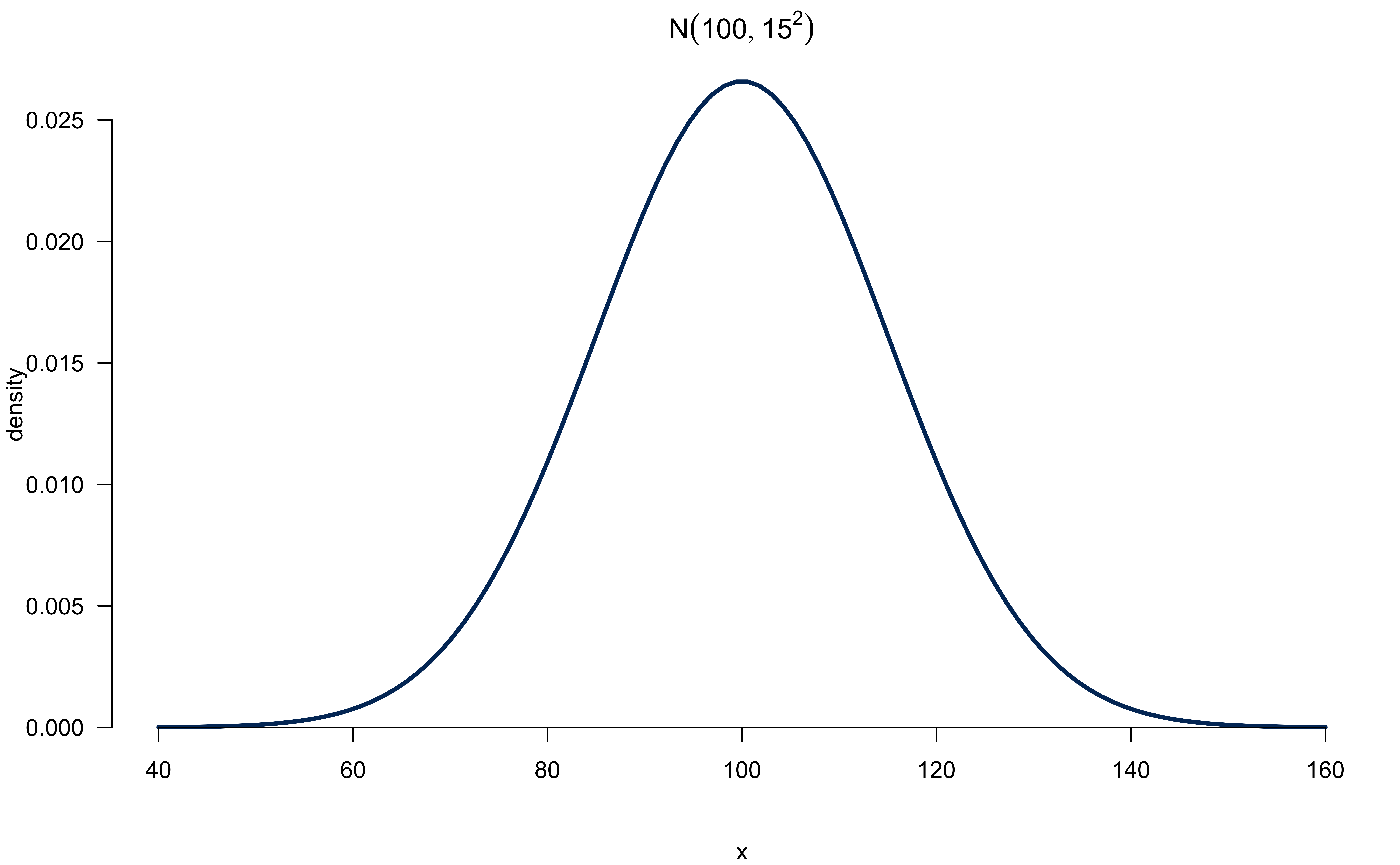
Draw Random Values from \(N(\mu, \sigma^2)\)
-
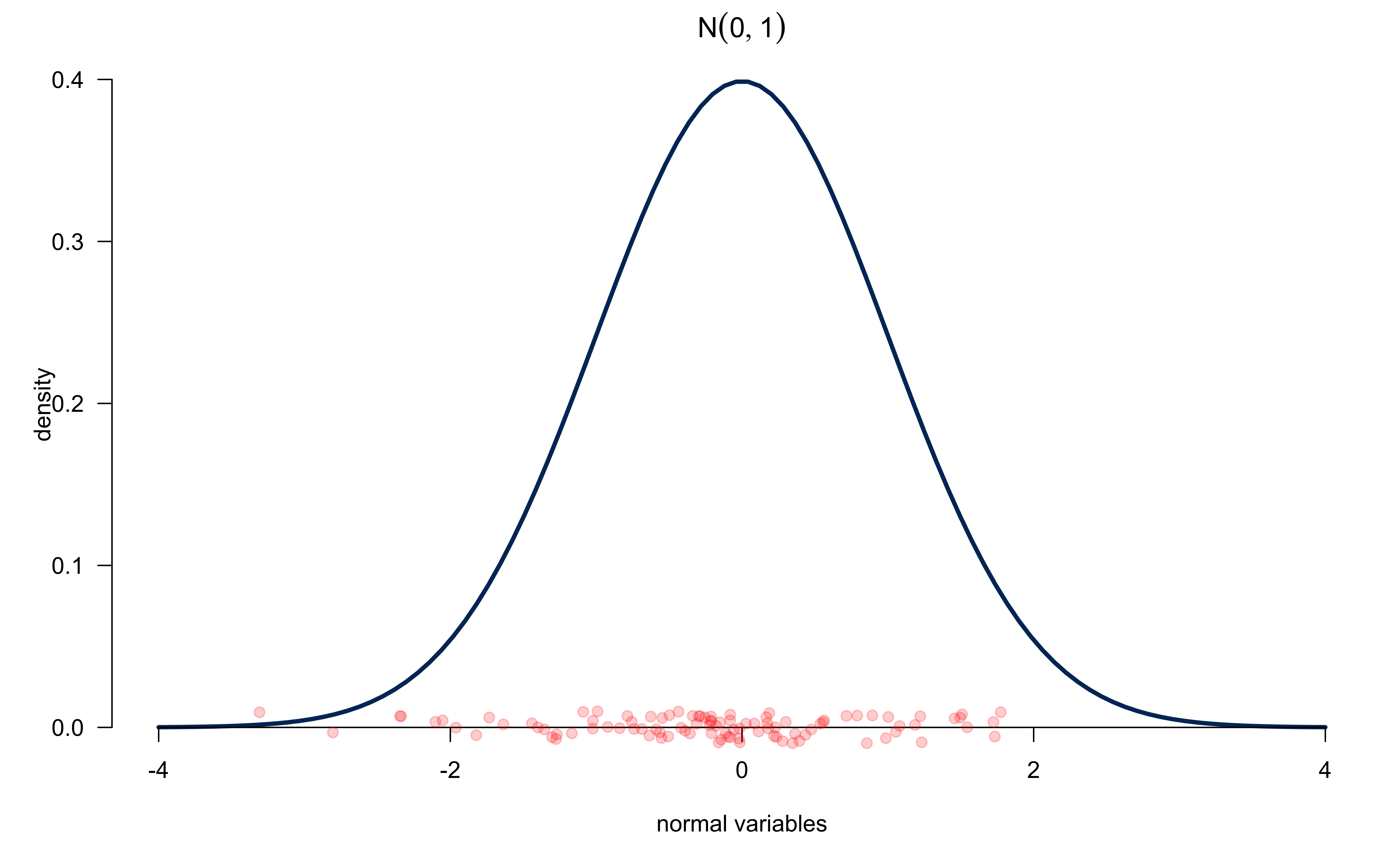
rnorm(n, mean, sd): Draw \(n\) observations from a normal distribution with meanmeanand standard deviationsd.
- \(100\) random draws from \(N(0, 1)\)
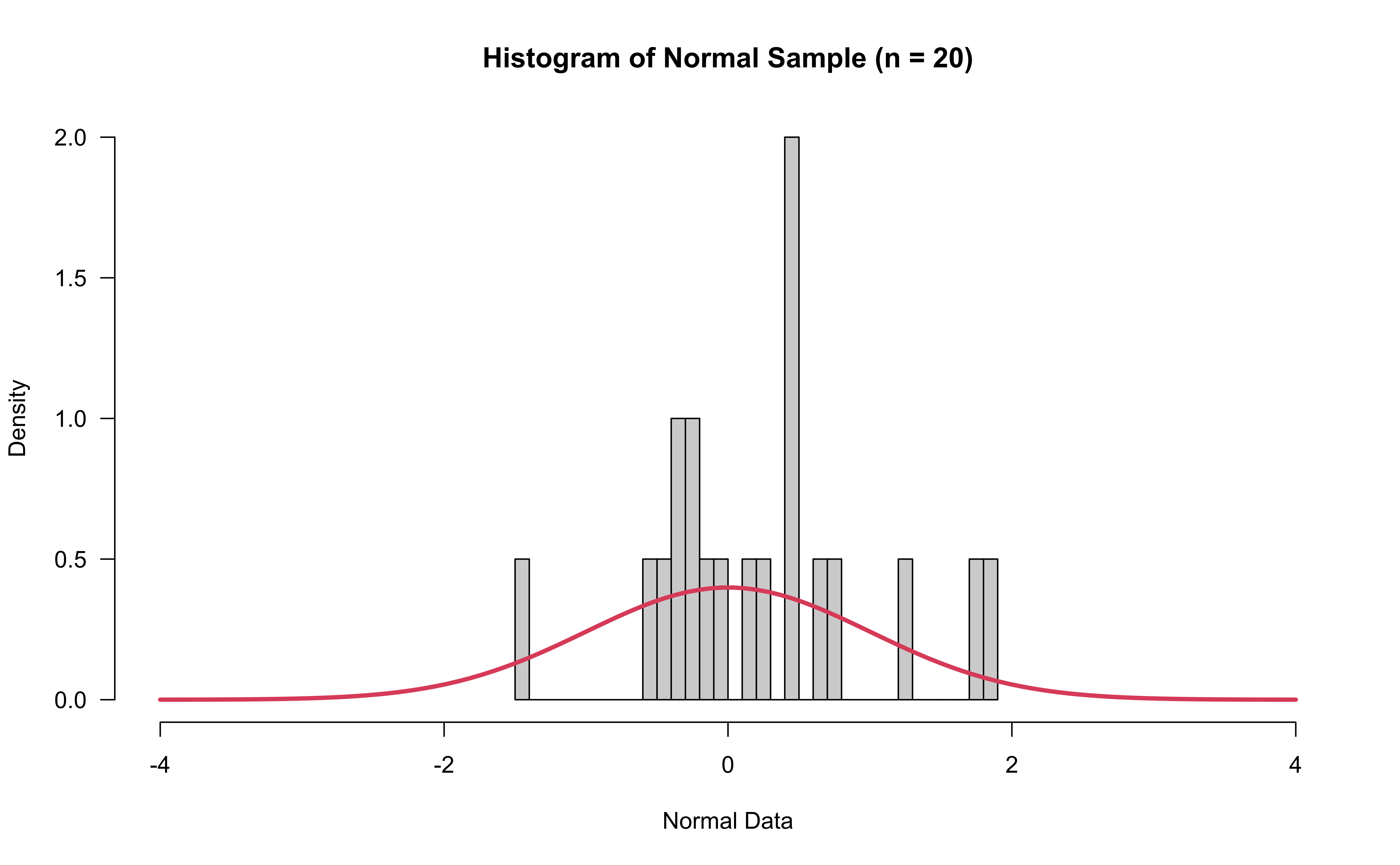
Histogram of Normal Data (n = 20)
Histogram of Normal Data (n = 200)
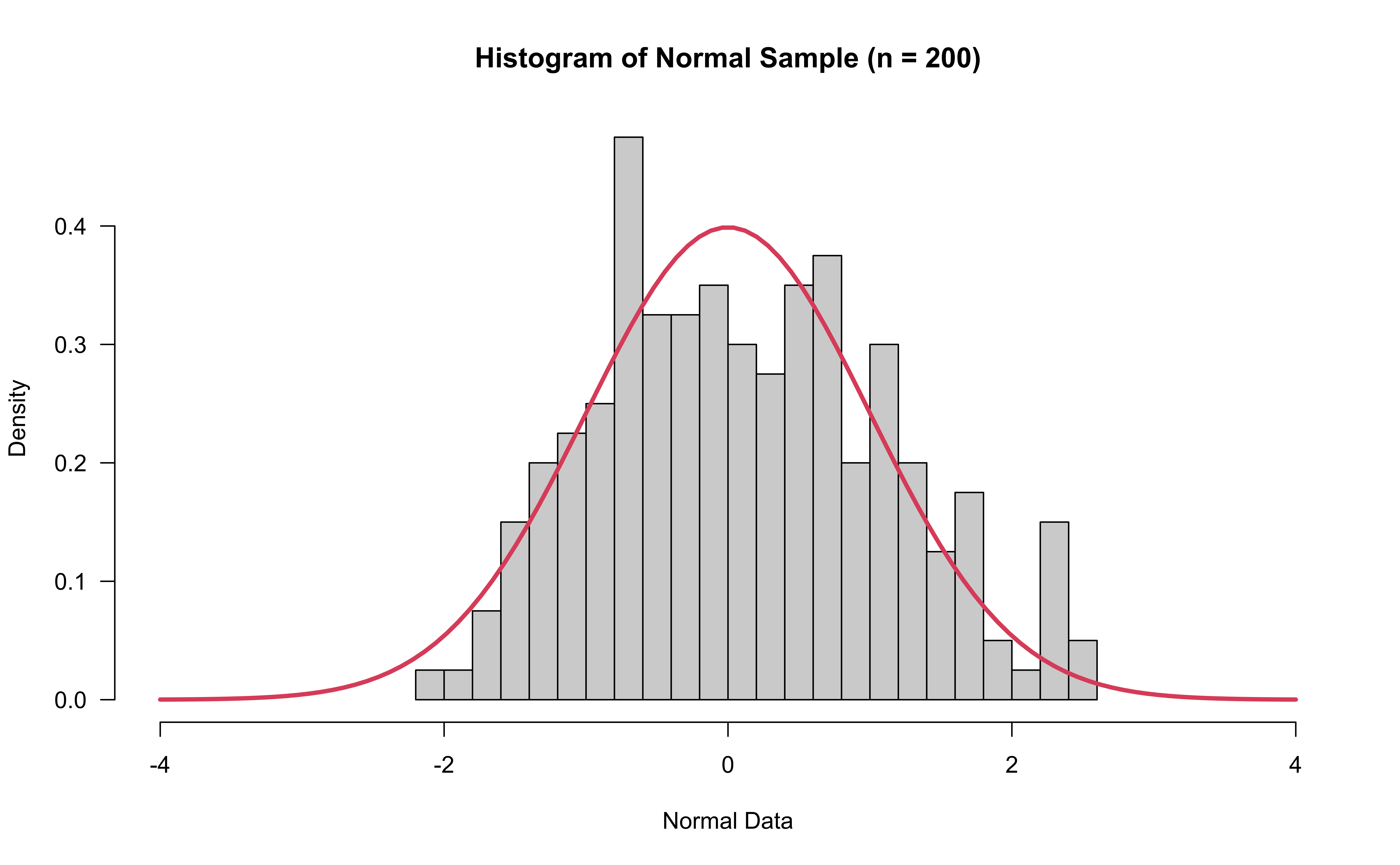
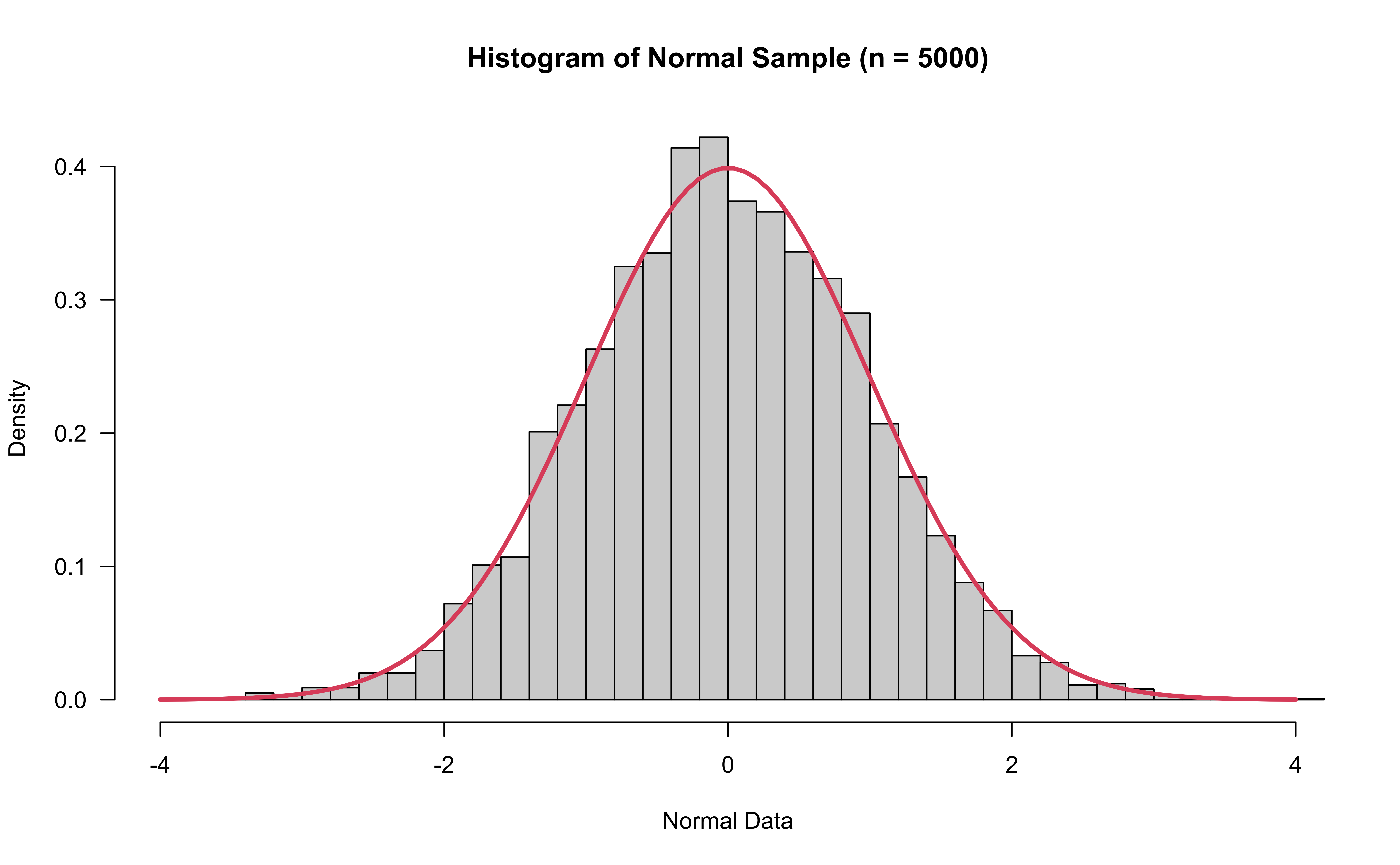
Histogram of Normal Data (n = 5000)
Compute Normal Probabilities
-
dnorm(x, mean, sd)to compute the density value \(f(x)\) (NOT probability) -
pnorm(q, mean, sd)to compute \(P(X \leq q)\) -
pnorm(q, mean, sd, lower.tail = FALSE)to compute \(P(X > q)\) -
pnorm(q2, mean, sd) - pnorm(q1, mean, sd)to compute \(P(q_1\leq X \leq q_2)\)
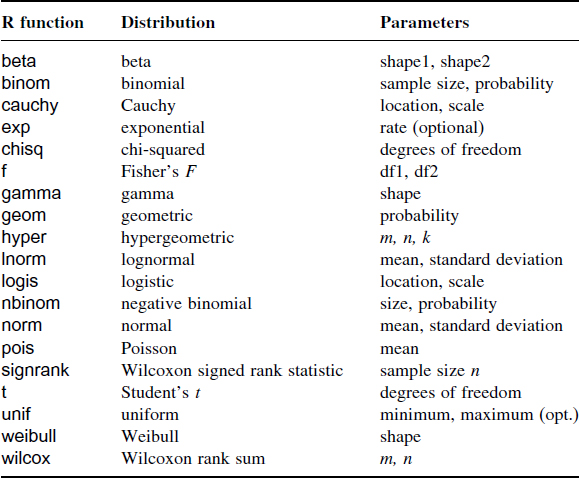
Distributions and Their R Function
Normal Curve
18-probability
In lab.qmd ## Lab 18 section,
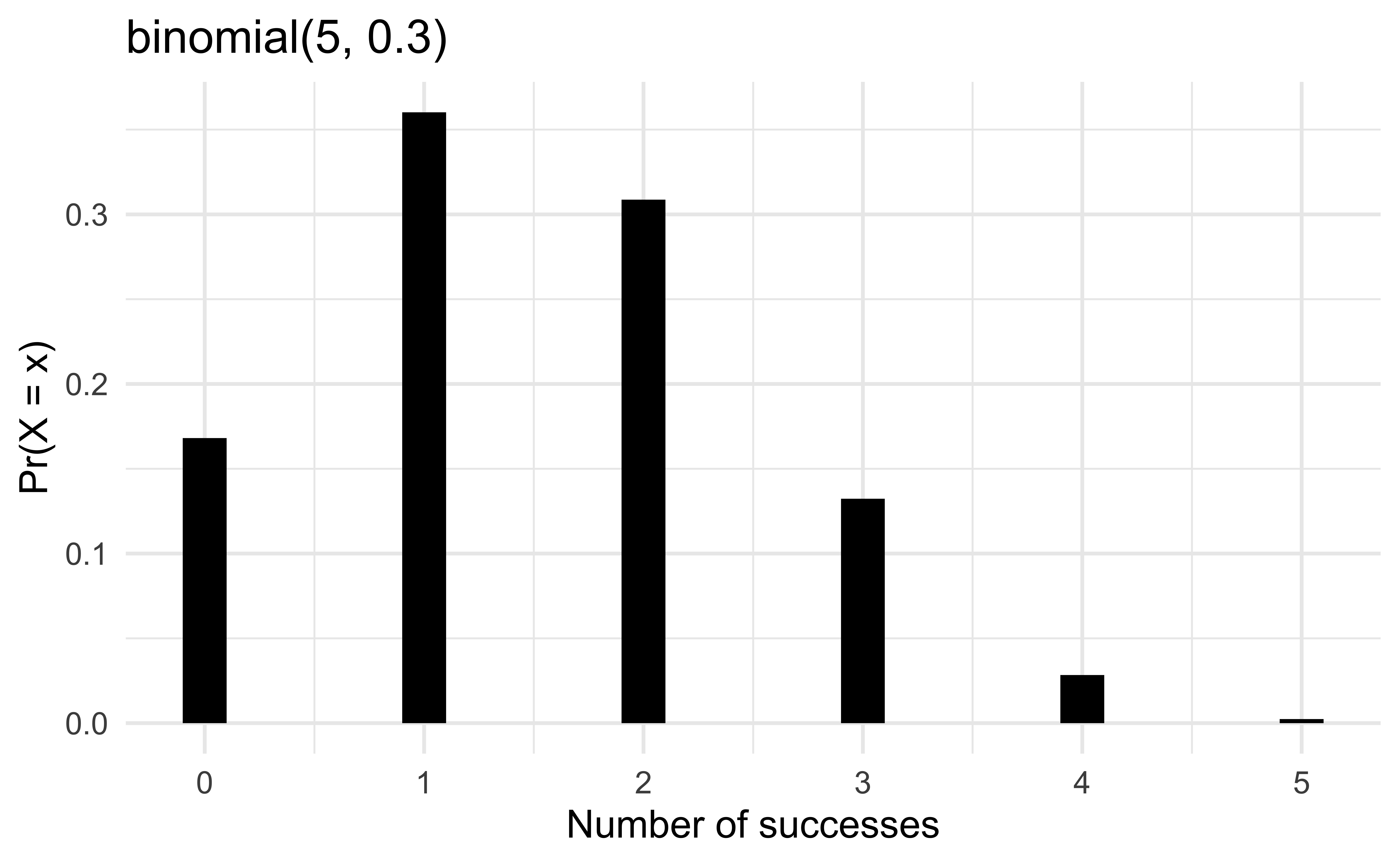
- Plot the probability function \(P(X = x)\) of \(X \sim \text{binomial}(n = 5, \pi = 0.3)\).
To use ggplot,
- Create a data frame saving all possible values of \(x\) and their corresponding probability using
dbinom(x, size = ___, prob = ___).
# A tibble: 6 × 2
x y
<int> <dbl>
1 0 0.168
2 1 0.360
3 2 0.309
4 3 0.132
5 4 0.0284
6 5 0.00243
2. Add geom_col()
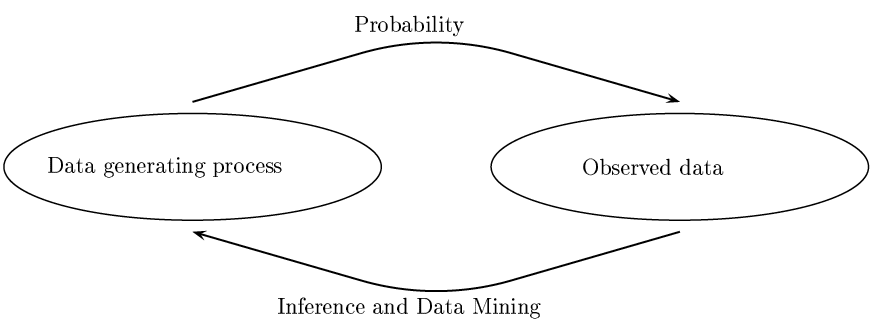
Probability vs. Statistics
- Probability : We know the process generating the data and are interested in properties of observations.
- Statistics : We observed the data (sample) and are interested in determining what is the process generating the data (population).
Terminology
Population (Data generating process): a group of subjects we are interested in studying
Sample (Data): a (representative) subset of our population of interest
Parameter: a unknown fixed numerical quantity derived from the population 1
Statistic: a numerical quantity derived from a sample
Common population parameters of interest and their corresponding sample statistic:
| Quantity | Parameter | Statistic (Point estimate) |
|---|---|---|
| Mean | \(\mu\) | \(\overline{x}\) |
| Variance | \(\sigma^2\) | \(s^2\) |
| Standard deviation | \(\sigma\) | \(s\) |
| proportion | \(p\) | \(\hat{p}\) |
\((1 - \alpha)100\%\) Confidence Interval for \(\mu\)
- With \(z_{\alpha/2}\) being \((1-\alpha/2)\) quantile of \(N(0, 1)\), \((1 - \alpha)100\%\) confidence interval for \(\mu\) is \[\left(\overline{X} - z_{\alpha/2} \frac{\sigma}{\sqrt{n}}, \,\, \overline{X} + z_{\alpha/2}\frac{\sigma}{\sqrt{n}}\right)\]
What if \(\sigma\) is unknown?
- \((1 - \alpha)100\%\) confidence interval for \(\mu\) becomes \[\left(\overline{X} - t_{\alpha/2, n-1} \frac{S}{\sqrt{n}},\,\, \overline{X} + t_{\alpha/2, n-1}\frac{S}{\sqrt{n}}\right),\] where \(t_{\alpha/2, n-1}\) is the \((1-\alpha/2)\) quantile of Student-t distribution with degrees of freedom \(n-1\).
Interpreting a 95% Confidence Interval
- We are 95% confident that blah blah blah . . .
If we were able to collect our sample data many times and build the corresponding confidence intervals, we would expect about 95% of those intervals would contain the true population parameter.
However,
We never know if in fact 95% of them do, or whether any particular interval contains the true parameter! 😱
❌ Cannot say “There is a 95% chance/probability that the true parameter is in the confidence interval.”
In practice we may only be able to collect one single data set.
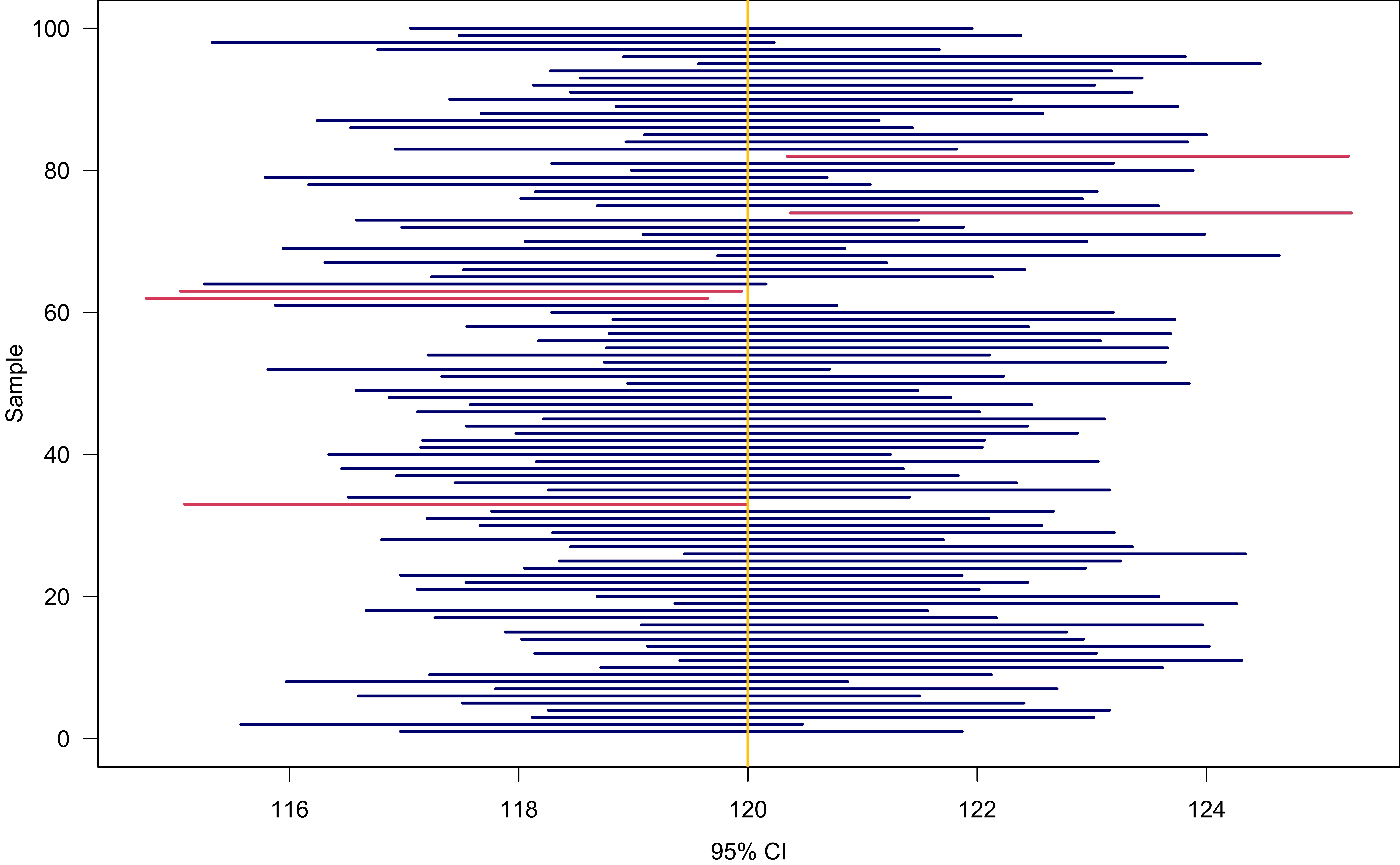
95% Confidence Interval Simulation
\(X_1, \dots, X_n \sim N(\mu, \sigma^2)\) where \(\mu = 120\) and \(\sigma = 5\).
Simulate 100 CIs for \(\mu\) when \(\sigma\) is known
Algorithm
- Generate 100 sampled data of size \(n\): \((x_1^1, x_2^1, \dots, x_n^1), \dots (x_1^{100}, x_2^{100}, \dots, x_n^{100})\), where \(x_i^m \sim N(\mu, \sigma^2)\).
- Obtain 100 sample means \((\overline{x}^1, \dots, \overline{x}^{100})\).
- For each \(m = 1, 2, \dots, 100\), compute the corresponding confidence interval \[\left(\overline{x}^m - z_{\alpha/2} \frac{\sigma}{\sqrt{n}}, \overline{x}^m + z_{\alpha/2}\frac{\sigma}{\sqrt{n}}\right)\]
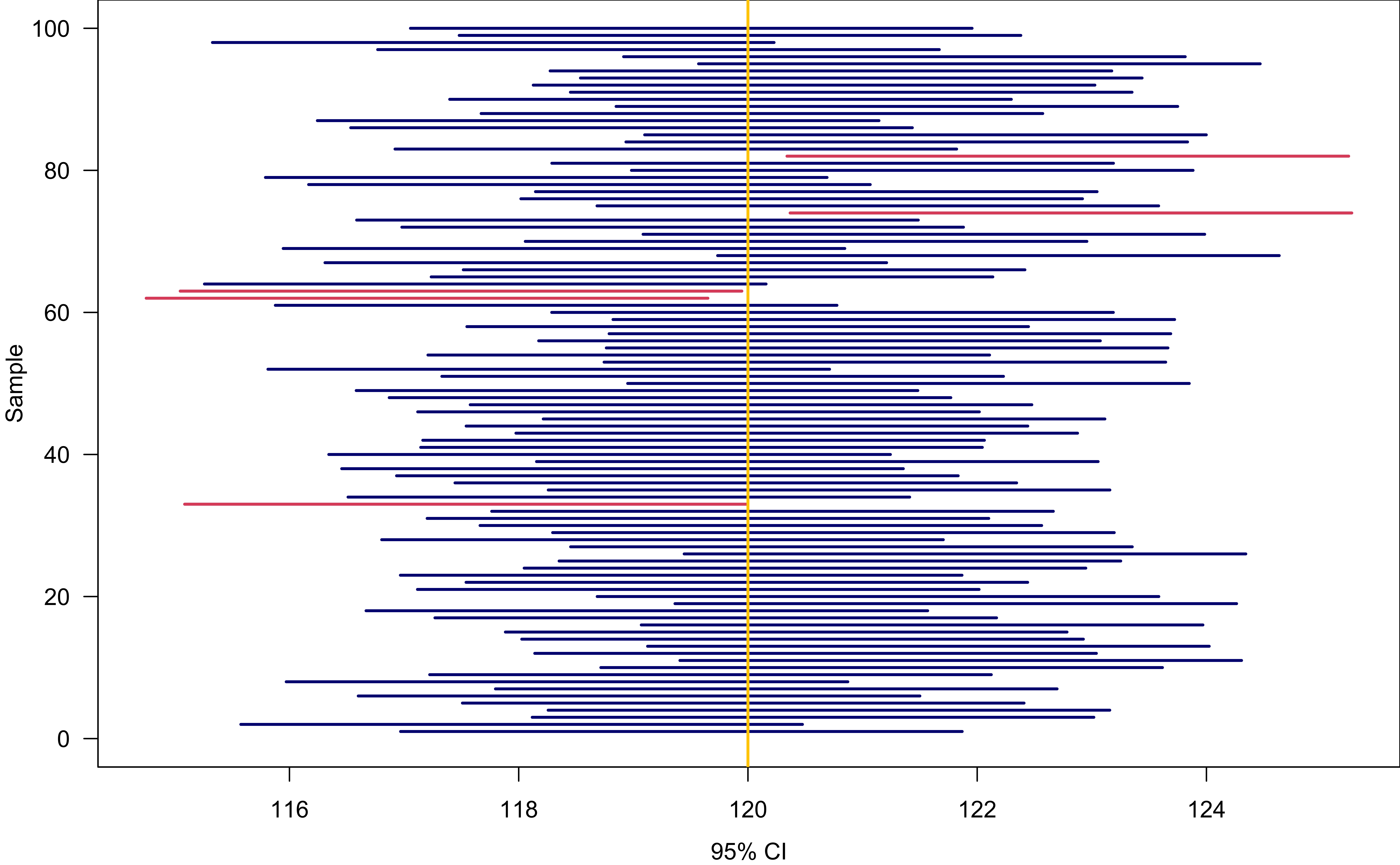
mu <- 120; sig <- 5
al <- 0.05; M <- 100; n <- 16
set.seed(2024)
x_rep <- replicate(M, rnorm(n, mu, sig))
xbar_rep <- apply(x_rep, 2, mean)
E <- qnorm(p = 1 - al / 2) * sig / sqrt(n)
ci_lwr <- xbar_rep - E
ci_upr <- xbar_rep + E
plot(NULL, xlim = range(c(ci_lwr, ci_upr)), ylim = c(0, 100),
xlab = "95% CI", ylab = "Sample", las = 1)
mu_out <- (mu < ci_lwr | mu > ci_upr)
segments(x0 = ci_lwr, y0 = 1:M, x1 = ci_upr, col = "navy", lwd = 2)
segments(x0 = ci_lwr[mu_out], y0 = (1:M)[mu_out], x1 = ci_upr[mu_out], col = 2, lwd = 2)
abline(v = mu, col = "#FFCC00", lwd = 2)
Random Generator
-
random.Generator.choice(): Generates a random sample from a given array
Normal Distribution from SciPy
Normal Curve Plotting