Data Splitting and K-Nearest Neighbors
MATH/COSC 3570 Introduction to Data Science
Department of Mathematical and Statistical Sciences
Marquette University
Training and Test Data
Prediction
- Goal: Build a good regression function or classifier in terms of prediction accuracy.
- The mechanics of prediction is easy:
- Plug in values of predictors to the model equation.
- Calculate the predicted value of the response \(\hat{y}\)
- Getting it right is hard! No guarantee that
- the model estimates are close to the truth
- your model performs as well with new data (test data) as it did with your sample data (training data)
Spending Our Data
- Several steps to create a useful model:
- Parameter estimation
- Model selection
- Performance assessment, etc.
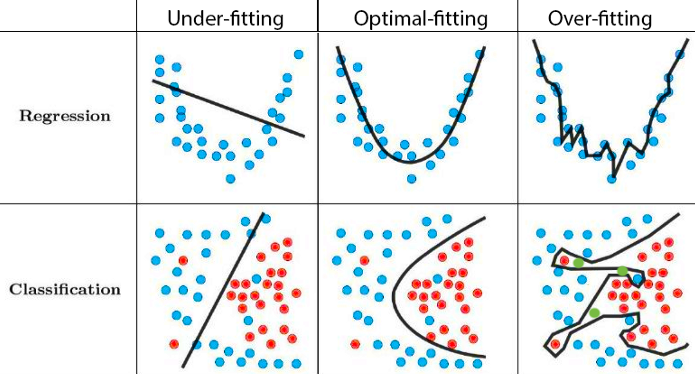
- Doing all of this using the entire data may lead to overfitting:
The model performs well on the training data, but awfully predicts the response on the new data we are interested.
Overfitting
The model performs well on the training data, but awfully predicts the response on the new data we are interested.
- Low error rate on observed data, but high prediction error rate on future unobserved data!
Splitting Data
Often, we don’t have another unused data to assess the performance of our model.
Solution: Pretend we have new data by splitting our data into training set and test set (validation set)!
-
Training set:
- Sandbox for model building
- Spend most of our time using the training set to develop the model
- Majority of the original sample data (75% - 80%)
-
Test set:
- Held in reserve to determine efficacy of one or two chosen models
- Critical to look at it once only, otherwise it becomes part of the modeling process
- Remainder of the data (20% - 25%)
initial_split() in ![rsample]()
body Data
What Makes a Good Classifier: Test Accuracy Rate
- The test accuracy rate associated with the test data \(\{x_j, y_j\}_{j=1}^J\): \[ \frac{1}{J}\sum_{j=1}^JI(y_j = \hat{y}_j),\] where \(\hat{y}_j\) is the predicted label resulting from applying the classifier to the test response \(y_j\) with predictor \(x_j\).
What is the value of \(J\) in our example?
- The best estimated classifier \(\hat{C}(x)\) trained from the training data for \(C(x)\) can be defined as the one producing the highest test accuracy rate or lowest test error rate.
K-Nearest Neighbors (KNN) Classifier
KNN classification uses majority voting:
Look for the most popular class label among its neighbors.
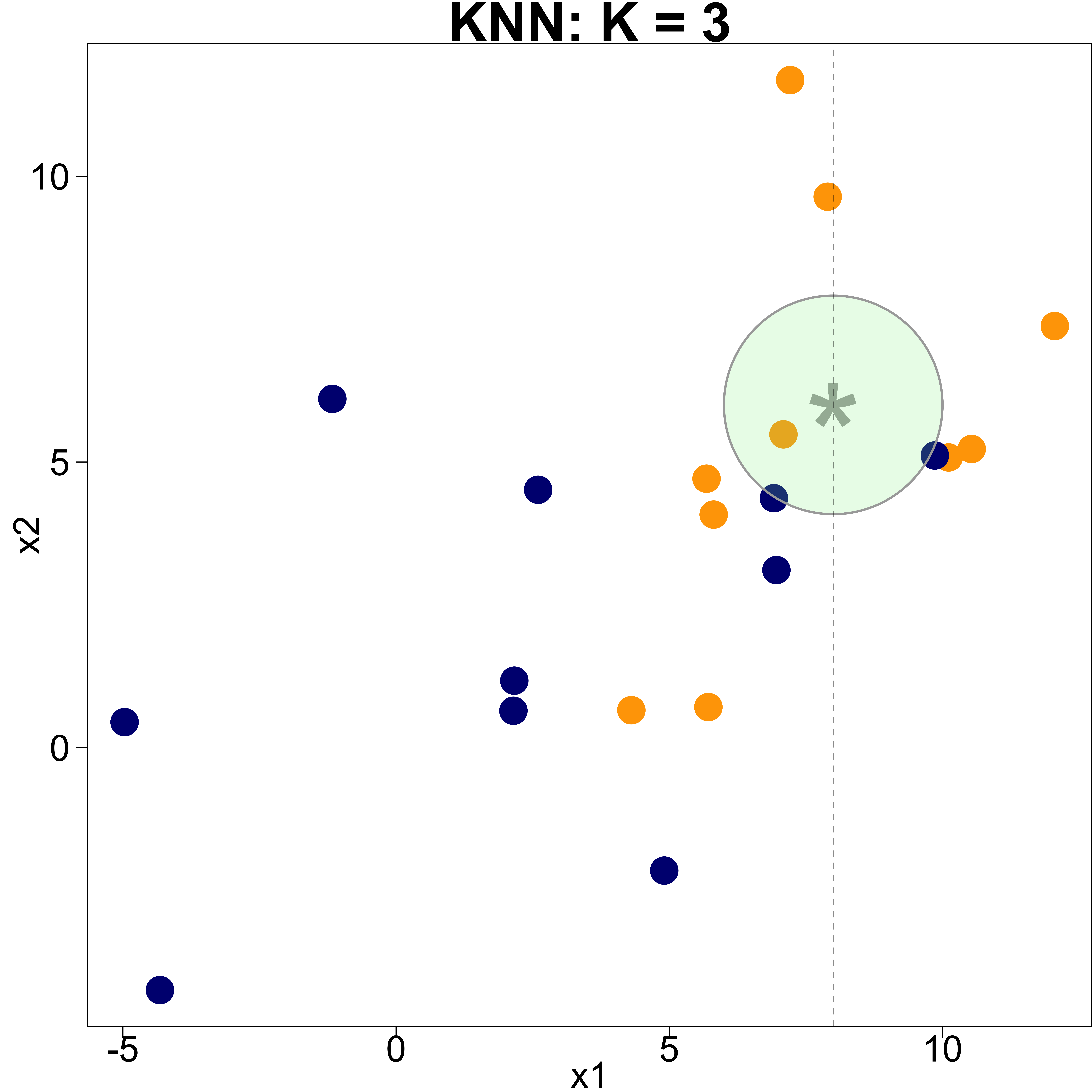
When predicting at \(x = (x_1, x_2) = (8, 6)\),
\[\begin{align} \hat{\pi}_{3Blue}(x = (8, 6)) &= \hat{P}(Y = \text{Blue} \mid x = (8, 6))\\ &= \frac{2}{3} \end{align}\]
\[\begin{align} \hat{\pi}_{3Orange}(x = (8, 6)) &= \hat{P}(Y = \text{Orange} \mid x = (8, 6))\\ &= \frac{1}{3} \end{align}\]
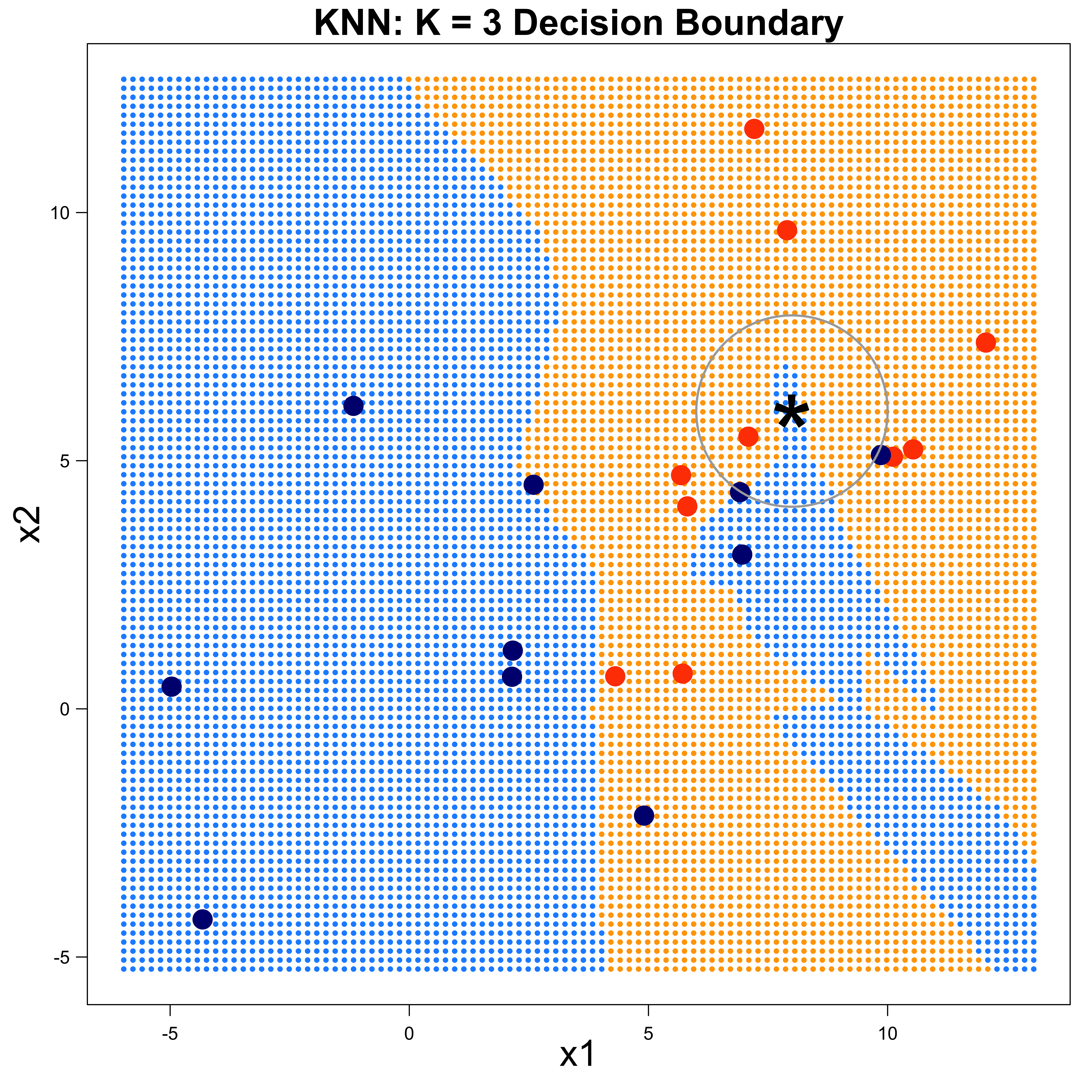
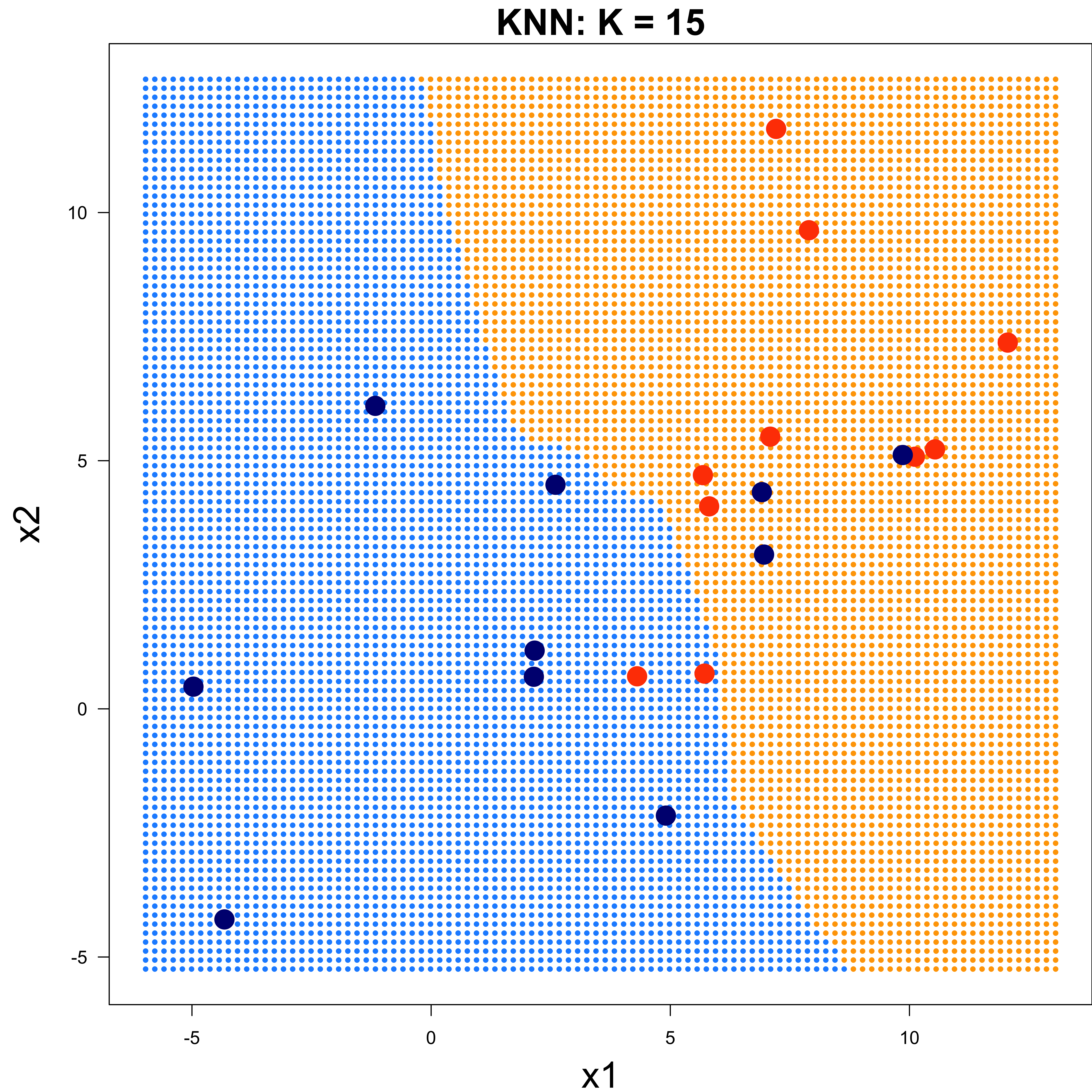
KNN Decision Boundary
Blue grid indicates the region in which a test response is assigned to the blue class.
We don’t know the true boundary (the true classification rule)!.
KNN Training ![recipes]()
-
Step 1: Create recipe:
recipes::recipe()
Standardize predictors before doing KNN!
KNN Training ![parsnip]()
-
Step 2: Specify Model:
parsnip::nearest_neighbor()
KNN Training ![workflows]()
-
Step 3: Fitting by creating workflow:
workflows::workflow()
knn_out <-
workflows::workflow() |>
add_recipe(knn_recipe) |>
add_model(knn_mdl) |>
fit(data = df_trn)══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: nearest_neighbor()
── Preprocessor ────────────────────────────────────────────────────────────────
1 Recipe Step
• step_normalize()
── Model ───────────────────────────────────────────────────────────────────────
Call:
kknn::train.kknn(formula = ..y ~ ., data = data, ks = min_rows(3, data, 5))
Type of response variable: nominal
Minimal misclassification: 0.217
Best kernel: optimal
Best k: 3Prediction on Test Data
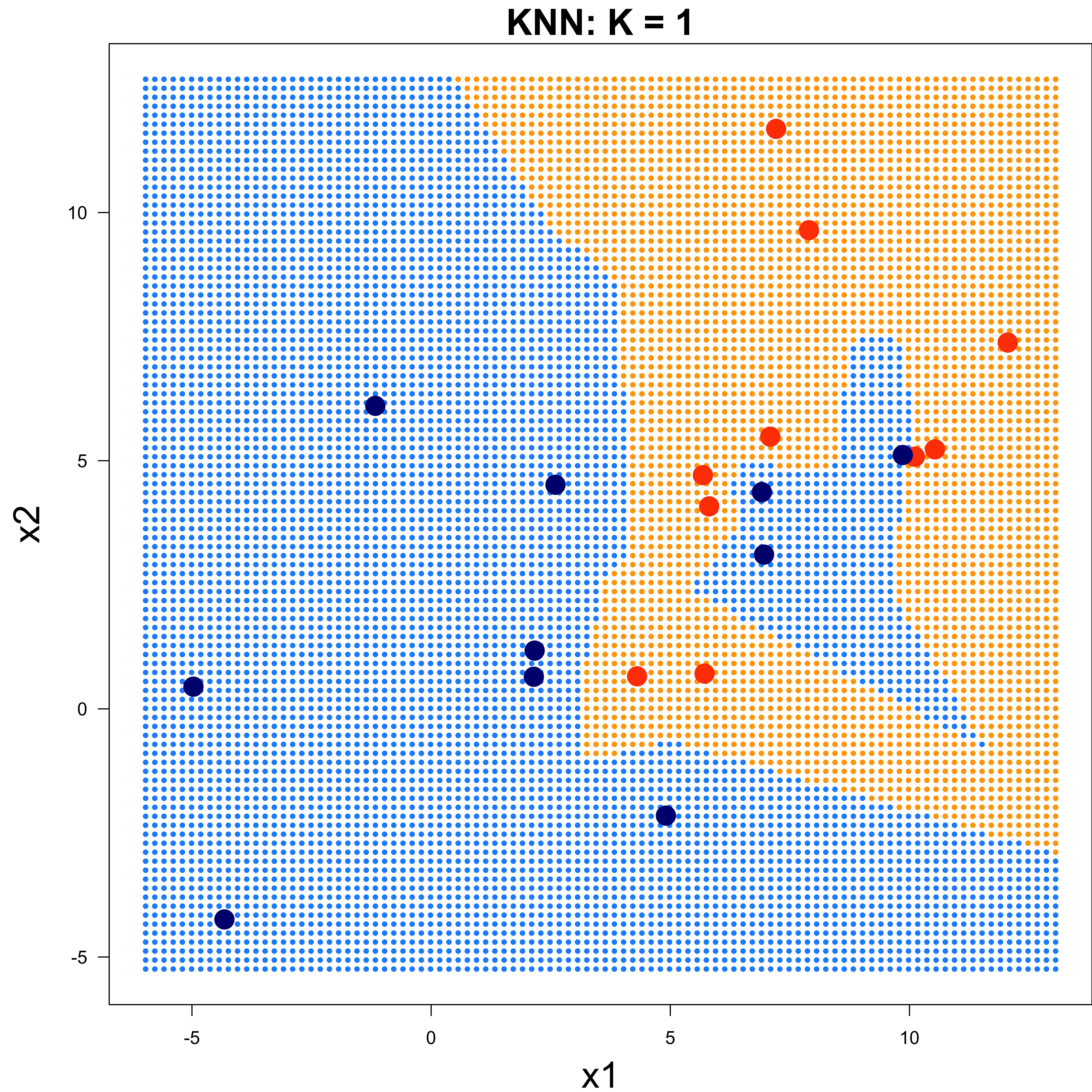
Which K Should We Use?
\(K\)-nearest neighbors has no model parameters, but a tuning parameter \(K\).
This is a parameter which determines how the model is trained, not a parameter that is learned through training.
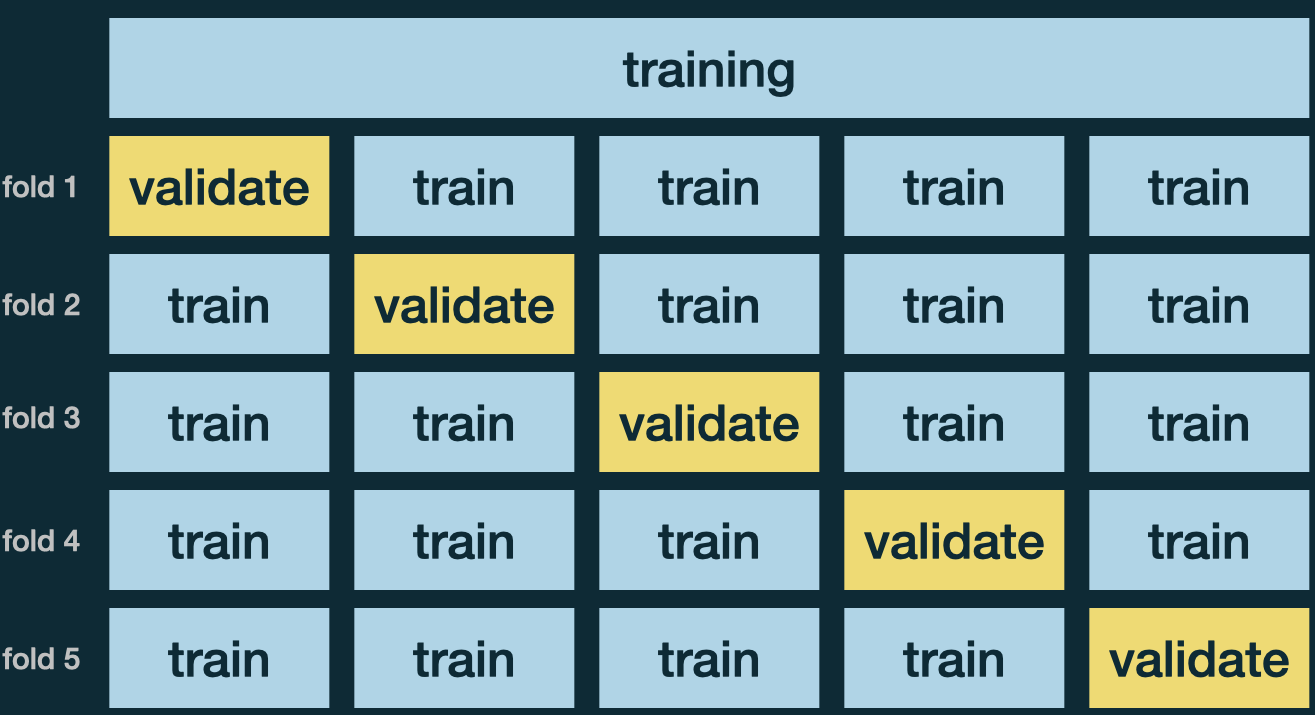
\(v\)-fold Cross Validation
Use \(v\)-fold Cross Validation (CV) to choose tuning parameters. (MATH 4750 Computational Statistics)
Usually use \(v = 5\) or \(10\).
- IDEA:
- Prepare \(v\) CV data sets
- Create a sequence of values of \(K\)
- For each value of \(K\), run CV, and obtain an accuracy rate
- Choose the \(K\) with the highest accuracy rate
Final Model
Final Model Performance
sklearn.neighbors
sklearn.model_selection
sklearn.model_selection.train_test_split
sklearn.neighbors.KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors = 3)
X_trn = np.array(X_trn)
X_tst = np.array(X_tst)
knn.fit(X_trn, y_trn)KNeighborsClassifier(n_neighbors=3)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
KNeighborsClassifier(n_neighbors=3)
Prediction
array([[22, 5],
[12, 21]])22-K Nearest Neighbors
In lab.qmd ## Lab 22 section,
use
HEIGHTandWAISTto predictGENDERusing KNN with \(K = 3\).Generate the (test) confusion matrix.
Calculate (test) accuracy rate.
Does using more predictors predict better?
R Code
library(tidymodels)
## load data
bodydata <- read_csv("./data/body.csv")
body <- bodydata |>
select(GENDER, HEIGHT, WAIST) |>
mutate(GENDER = as.factor(GENDER))
## training and test data
set.seed(2024)
df_split <- initial_split(data = body, prop = 0.8)
df_trn <- training(df_split)
df_tst <- testing(df_split)
## KNN training
knn_recipe <- recipe(GENDER ~ HEIGHT + WAIST, data = df_trn) |>
step_normalize(all_predictors())
knn_mdl <- nearest_neighbor(neighbors = 3, mode = "classification")
knn_out <- workflow() |>
add_recipe(knn_recipe) |>
add_model(knn_mdl) |>
fit(data = df_trn)
## KNN prediction
knn_pred <- pull(predict(knn_out, df_tst))
table(knn_pred, df_tst$GENDER)
mean(knn_pred == df_tst$GENDER)Python Code
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
## load data
body = pd.read_csv('./data/body.csv')
X = body[['HEIGHT', 'WAIST']]
y = body['GENDER']
## training and test data
X_trn, X_tst, y_trn, y_tst = train_test_split(X, y, test_size=0.2, random_state=2024)
## KNN training
knn = KNeighborsClassifier(n_neighbors = 3)
X_trn = np.array(X_trn)
X_tst = np.array(X_tst)
knn.fit(X_trn, y_trn)
## KNN prediction
y_pred = knn.predict(X_tst)
from sklearn.metrics import confusion_matrix
confusion_matrix(y_tst, y_pred)
np.mean(y_tst == y_pred)


